Reverse Transcription Primer Design Tool
Brad Hook and Samantha Lewis
Promega Corporation
Publication Date: June 2017; tpub_185
Abstract
To initiate reverse transcription, reverse transcriptases require a short DNA oligonucleotide called a primer to bind the RNA template and serve as a starting point for synthesis of a new strand. Depending on the RNA template and the downstream applications, primers of three basic types are available: oligo(dT) primers, random primers and gene-specific primers. Each primer binds to a complementary sequence on the RNA template to facilitate reverse transcription, the difference between these options is the sequence that they bind. Gene specific primers bind target sequences contained within a single mRNA of interest and only that region is amplified; these primers are often used for one-step RT-qPCR reactions. Oligo(dT) primers amplify only mRNAs containing a poly(A) tail, since that is where the primer binds to promote reverse transcription. Random primers amplify most RNA species, including degraded RNA and viral genomes. In some cases both oligo(dT) and random primers can be used to combine the benefits of both primer types. We compared the GoScript™ Reverse Transcription Mixes, Oligo(dT) and Random Primers, to a 50/50 mixture of the reagents for ability to reverse transcribe three RNA transcripts.
Introduction
Reverse transcriptases (RTs), also known as RNA-directed DNA polymerases, were first identified in retroviruses (1) (2) . In addition to the critical role these enzymes play in a variety of major human diseases (HIV, hepatitis, Zika and some cancers), RTs are a fundamental component of the molecular biologist's toolbox. RT activity is critical for many basic techniques including real-time and endpoint RT-PCR, RNA-Seq, labeled-probe generation for microarrays and cDNA cloning. The ideal reverse transcriptase is robust (highly active under a variety of conditions) and converts all primed RNA within a sample to cDNA, regardless of its abundance, length or secondary structure.
GoScript™ Reverse Transcription Mixes are convenient reagents that include an optimized set of components designed for efficient first-strand cDNA synthesis. Priming using random primers is the most general method of initiating cDNA synthesis from a variety of RNA templates. Random primers can be used to prime first-strand cDNA synthesis from all RNA molecules, including those that do not possess a poly(A)+ tail and RNA isolated from prokaryotic sources. Oligo(dT) priming initiates first-strand synthesis by annealing to the 3′ end of any polyadenylated RNA molecule. Reverse transcription of total RNA or mRNA will yield oligo(dT)-primed products from any poly(A)+ RNA.
When choosing between oligo(dT) and random primers, the targets of interest and primer location should be considered. Oligo(dT) primers should not be used for amplification of RNA species without poly(A) tails but it can be very effective with full length RNA transcription, keeping in mind that some 3' bias can occur with certain targets. Alternatively, while random primers can be used with most RNA species, some very long mRNAs may not be completely reverse transcribed when primed with random primers using standard conditions, resulting in truncated cDNAs. Using a mixture of oligo(dT) and random primers can offer the strengths of both without their pitfalls. Ultimately, the best way to determine which primer to use is by understanding your target of interest and, when in doubt, experimental testing.
We performed a comparison of the GoScript™ Reverse Transcription Mix, Oligo(dT), GoScript™ Reverse Transcription Mix, Random Primers, and a 50/50 mixture of the two mixes (containing both random primers and oligo(dT) primers). We compared these RT mixes for their utility in detecting three transcripts by real-time PCR. The transcripts ranged from high to low cellular abundance (3) .
GoScript™ Reverse Transcription Mix, Oligo(dT), Cat.# A2790
GoScript™ Reverse Transcription Mix, Random Primers, Cat.# A2800
GoTaq® qPCR Master Mix , Cat.# A6001
Universal Human Reference RNA, Agilent, Cat.# 740000
CFX96™ Real-Time Detection System, BioRad
Veriti™ 96-Well Thermal Cycler, Applied Biosystems
Protocol
An 8-point standard curve of Universal Human Reference RNA was made in nuclease-free water with 10-fold dilutions from 100ng/µl down to 1fg/µl. Reverse transcription was performed according to the GoScript™ Reverse Transcription Mix Product Information protocols (Literature # 9PIA279 for Oligo(dT) and # 9PIA280 for Random Primers) . For the mixed protocol, instead of 4µl of a single mix, 2µl per reaction of GoScript™ Reverse Transcription Mix, Oligo(dT), and 2µl per reaction of GoScript™ Reverse Transcription Mix, Random Primers, was used.
Real-Time qPCR was performed with 2µl of reverse transcriptions in a 20µl total reaction volume using GoTaq® qPCR Master Mix. Three different targets were examined, GAPDH, SDHA and UBC, which are predicted to be present at high, medium, and low expression levels, respectively. Primer sequences are listed below and were obtained from a reference by Cicinnati et al. 2008 (3) .
GAPDH: Forward 5´-TGCACCACCAACTGCTTAGC-3´, Reverse 5´-GGCATGGACTGTGGTCATGA-3´
SDHA: Forward 5´-TGGGAACAAGAGGGCATCTG-3´, Reverse 5´-CCACCACTGCATCAAATTCATG-3´
UBC: Forward 5´-CGGTGAACGCCGATGATTAT-3´, Reverse 5´-ATCTGCATTGTCAAGTGACGA-3´
Results
Reverse transcription was performed on a serial dilution of RNA using three mixes: GoScript™ Reverse Transcription Mix, Oligo(dT), GoScript™ Reverse Transcription Mix, Random Primers and a 50/50 mixture of the two mixes. Following reverse transcription, qPCR was performed with each sample using 2µl of input. Expression of three genes was examined by qPCR; these genes had varying expression levels with GAPDH being highest and UBC being the lowest expressed. Figure 1 shows the Cq values of each replicate for the three genes tested. Similar Cq values were seen for random primers and mixed conditions, while oligo(dT) had, on average, higher Cq values for amplifications with SDHA and UBC primers. This highlights the importance of optimizing the reverse transcription protocol for the specific primer set of interest when optimal results are required. Cq values with averages from each primer set and condition are shown in Table 2. No significant differences in slope, linearity or efficiency were observed with any of the mixes tested despite differences in Cq values (Table 3).
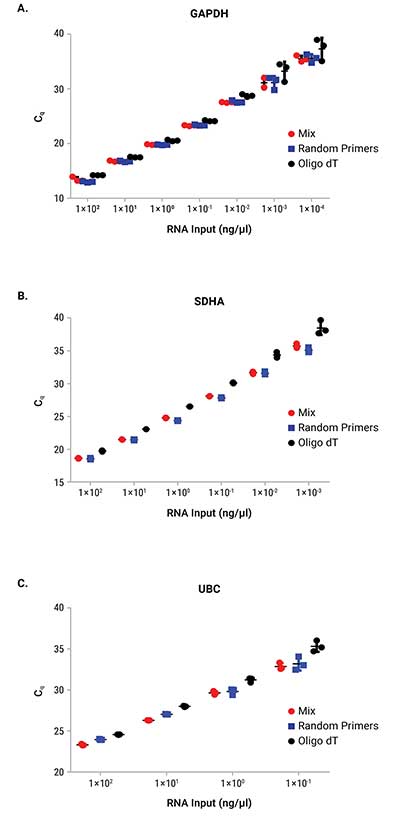 Figure 1. Cq values from qPCR with RNA that was reverse transcribed with GoScript™ Reverse Transcription Mixes.
Figure 1. Cq values from qPCR with RNA that was reverse transcribed with GoScript™ Reverse Transcription Mixes.
Three replicates for each reaction volume are shown with error bars representing standard deviation from the mean. Panel A. GAPDH. Panel B. SDHA. Panel C. UBC.
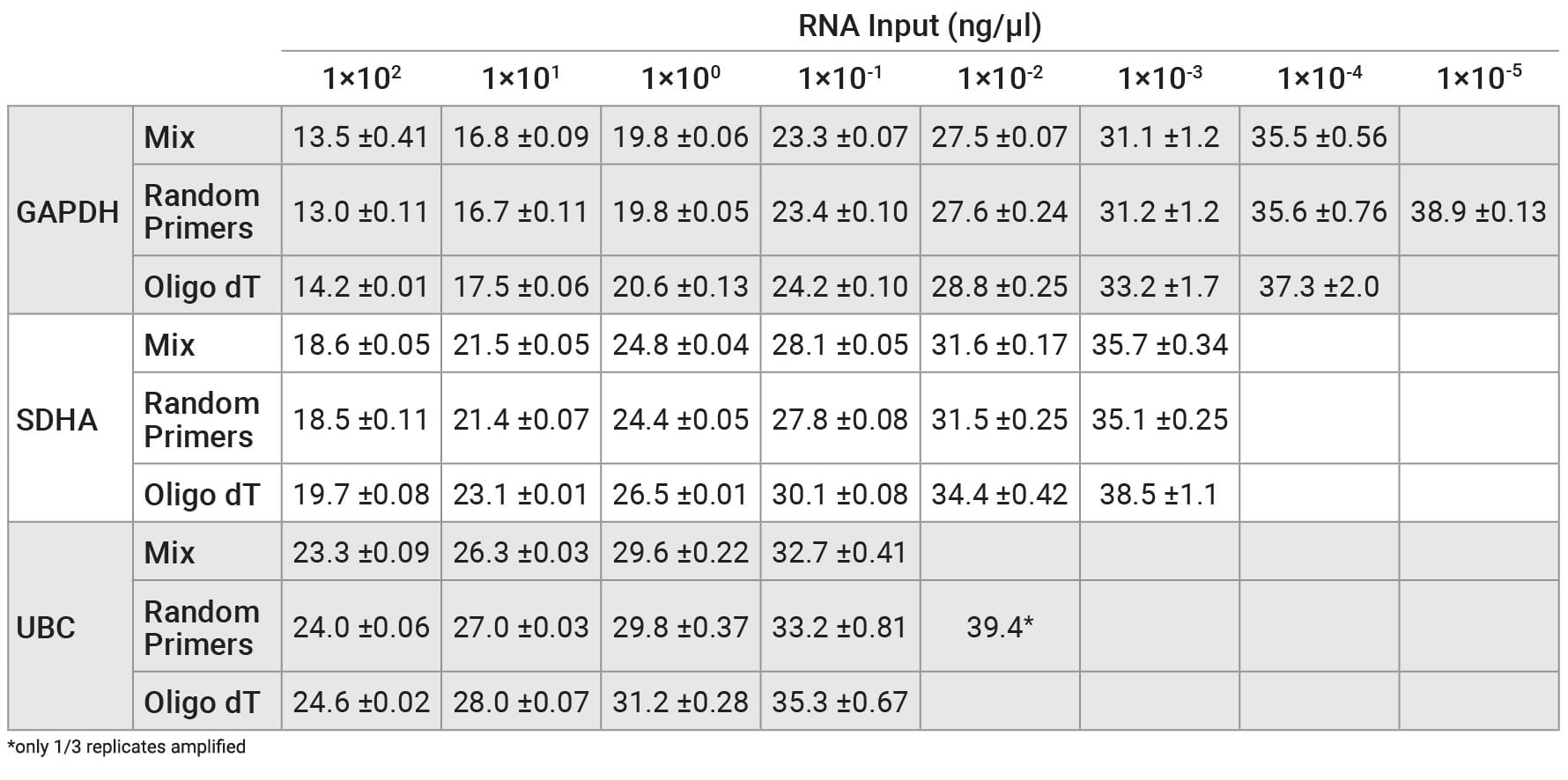 Table 1. Average Cq values from qPCR with RNA that was reverse transcribed using GoScript™ Reverse Transcription Mix, Oligo(dT), GoScript™ Reverse Transcription Mix, Random Primers, and 50/50 mixture of reagents.
Table 1. Average Cq values from qPCR with RNA that was reverse transcribed using GoScript™ Reverse Transcription Mix, Oligo(dT), GoScript™ Reverse Transcription Mix, Random Primers, and 50/50 mixture of reagents.
Cq values are shown as averages ± standard deviation of n=3.
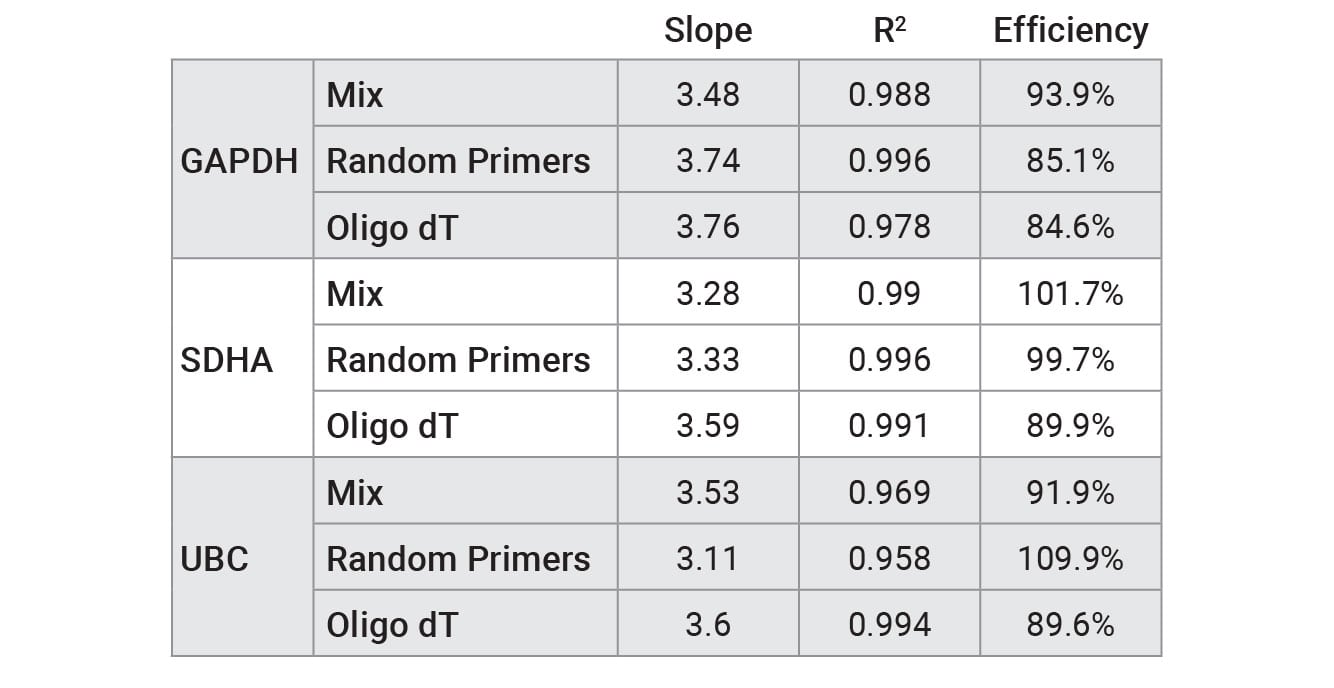 Table 2. Performance of qPCR primer sets tested with RNA that was reverse transcribed using GoScript™ Reverse Transcription Mix, Oligo(dT), GoScript™ Reverse Transcription Mix, Random Primers, and 50/50 mixture of reagents.
Table 2. Performance of qPCR primer sets tested with RNA that was reverse transcribed using GoScript™ Reverse Transcription Mix, Oligo(dT), GoScript™ Reverse Transcription Mix, Random Primers, and 50/50 mixture of reagents.
Values were calculated by plotting Cq versus RNA concentration for standard curve and using a linear curve fit model.
The ideal efficiency is 100% (slope = –3.3), but an efficiency in the range of 90–110% is acceptable according to the MIQE guidelines (4) . Further primer optimization is required to increase some of the efficiencies to the 90–110% range.
Summary
A mixture of GoScript™ Reverse Transcription Mix, Oligo(dT), and GoScript™ Reverse Transcription Mix, Random Primers, can be used for reverse transcription with no significant difference in qPCR performance. For optimal results when designing an assay, oligo(dT) and random primers should both be tested to determine which will provide the best results. If desired, a mix of the two can also be used.
How to Cite This Article
Scientific Style and Format, 7th edition, 2006
Hook, B. and Lewis, S. Primer selection guide for use with GoScript® Reverse Transcription Mixes. [Internet] June 2017; tpub_185. [cited: year, month, date]. Available from: https://worldwide.promega.com/resources/pubhub/primer-selection-guide-for-use-with-goscript-reverse-transcription-mixes/
American Medical Association, Manual of Style, 10th edition, 2007
Hook, B. and Lewis, S. Primer selection guide for use with GoScript® Reverse Transcription Mixes. Promega Corporation Web site. https://worldwide.promega.com/resources/pubhub/primer-selection-guide-for-use-with-goscript-reverse-transcription-mixes/ Updated June 2017; tpub_185. Accessed Month Day, Year.
Reverse Transcription Primer Design Tool
Source: https://www.promega.com/resources/pubhub/primer-selection-guide-for-use-with-goscript-reverse-transcription-mixes/
Posted by: thomasreackagots.blogspot.com

0 Response to "Reverse Transcription Primer Design Tool"
Post a Comment